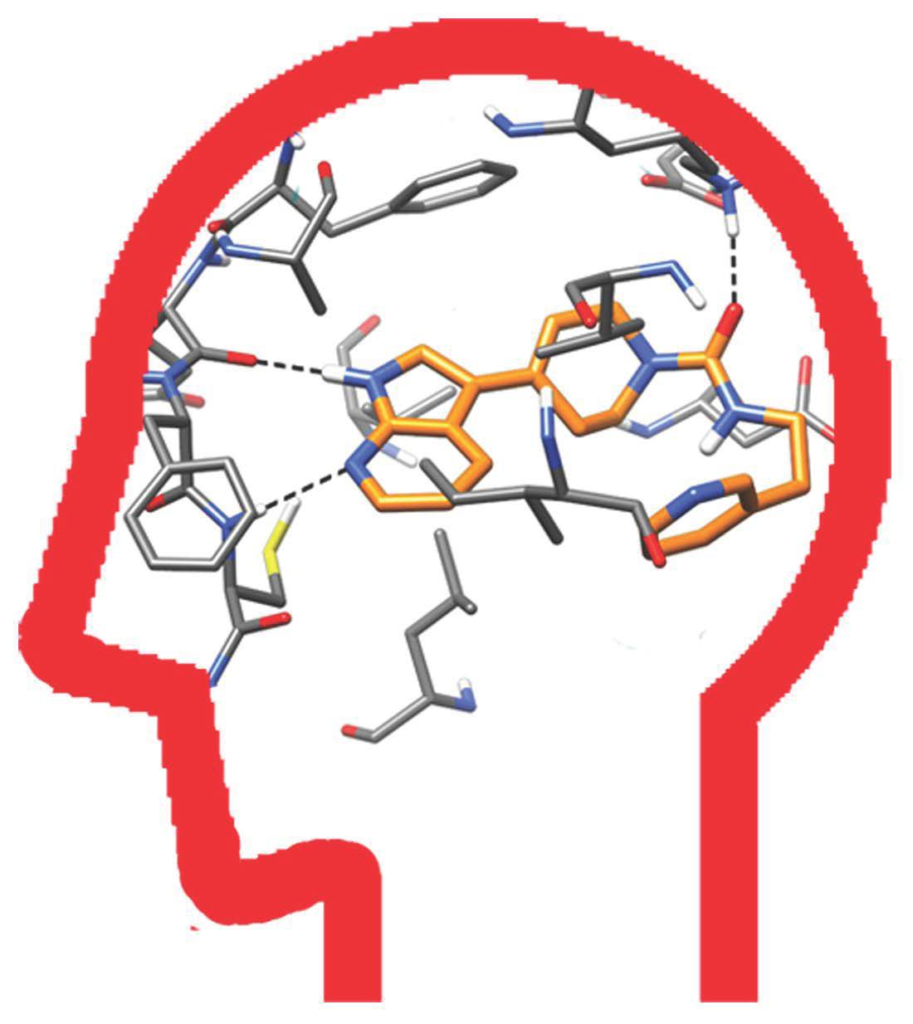
Protein kinases represent one of the largest groups of proteins in the human genome and they play a major role in eukaryotic signal transduction via regulation of the phosphorylation states and thus the cellular functions of substrate proteins.
Part of our team is dedicated to develop new potent kinase modulators through computational studies, chemical synthesis and in vitro assays; thus providing hit identification and lead optimization and in silico screenig platforms.
Publications:
204.
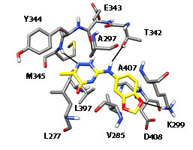
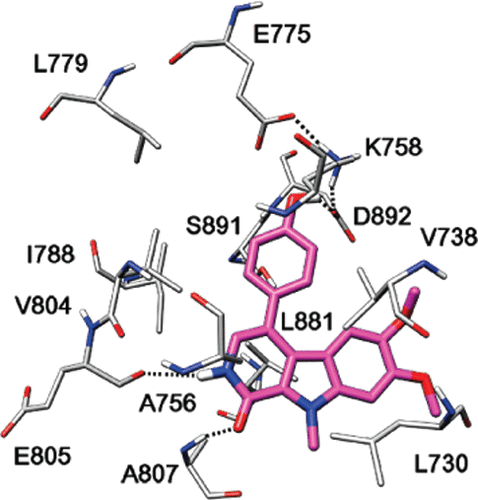
Machine Learning-Based Virtual Screening for the Identification of Cdk5 Inhibitors. Di Stefano M, Galati S, Ortore G, Caligiuri I, Rizzolio F, Ceni C, Bertini S, Bononi G, Granchi C, Macchia M, Poli G, Tuccinardi T. Int J Mol Sci. 2022, 23(18):10653.
169.
Three-Dimensional Interactions Analysis of the Anticancer Target c-Src Kinase with Its Inhibitors. Jha V, Macchia M, Tuccinardi T, Poli G. Cancers (Basel). 2020, 12(8):2327.
120.
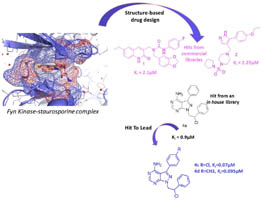
Binding investigation and preliminary optimisation of the 3-amino-1,2,4-triazin-5(2H)-one core for the development of new Fyn inhibitors. Poli G, Lapillo M, Granchi C, Caciolla J, Mouawad N, Caligiuri I, Rizzolio F, Langer T, Minutolo F, Tuccinardi T. J Enzyme Inhib Med Chem. 2018, 33(1):956-961.
87.
Studies on the ATP binding site of Fyn kinase for the identification of new inhibitors and their evaluation as potential agents against tauopathies and tumors. Tintori C, La Sala G, Vignaroli G, Botta L, Fallacara AL, Falchi F, Radi M, Zamperini C, Dreassi E, Dello Iacono L, Orioli D, Biamonti G, Garbelli M, Lossani A, Gasparrini F, Tuccinardi T, Laurenzana I, Angelucci A, Maga G, Schenone S, Brullo C, Musumeci F, Desogus A, Crespan E, Botta M. J Med Chem. 2015 58(11):4590-4609.
78.
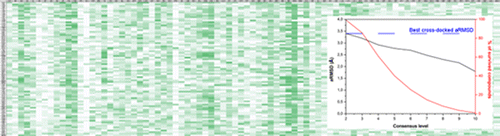
Extensive Consensus Docking Evaluation for Ligand Pose Prediction and Virtual Screening Studies. Tuccinardi T, Poli G, Romboli V, Giordano A, Martinelli A. J Chem Inf Model. 2014, 54(10):2980-2986.
77.
Computational approaches for the identification and optimization of Src family kinases inhibitors. Poli G, Martinelli A, Tuccinardi T. Curr Med Chem. 2014, 21(28):3281-3293.
69.
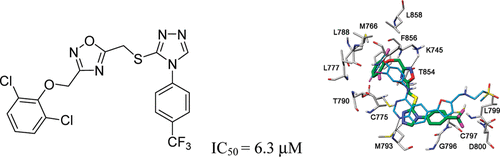
Identification of new FYN kinase inhibitors using a FLAP-based approach. Poli G, Tuccinardi T, Rizzolio F, Caligiuri I, Botta L, Granchi C, Ortore G, Minutolo F, Schenone S, Martinelli A. J Chem Inf Model. 2013, 53(10):2538-2547.
56.
Protein Kinase Homology Models: Recent Developments and Results. Tuccinardi T, Martinelli A. Curr Med Chem. 2011;18(19):2848-2853.
45.
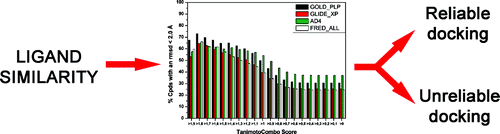
Protein Kinases: Docking and Homology Modeling Reliability. Tuccinardi T, Botta M, Giordano A, Martinelli A. J Chem Inf Model. 2010, 50(8):1432-1441.
44.
CDK inhibitors: from the bench to clinical trials. Rizzolio F, Tuccinardi T, Caligiuri I, Lucchetti C, Giordano A. Curr. Drug Targets. 2010;11(3):279-290.
36.
Computational Studies of Epidermal Growth Factor Receptor: Docking Reliability, Three-Dimensional Quantitative Structure−Activity Relationship Analysis, and Virtual Screening Studies. La Motta C, Sartini S, Tuccinardi T, Nerini E, Da Settimo F, Martinelli A. J Med Chem 2009, 52(4):964-975.
33.
Synthesis, modeling and RET protein kinase inhibitory activity of 3- and 4-substituted-β-carbolin-1-ones. Cincinelli R, Cassinelli G, Dallavalle S, Lanzi C, Merlini L, Botta M, Tuccinardi T, Martinelli A, Penco S, Zunino F. J Med Chem 2008, 51(24):7777-7787.
19.
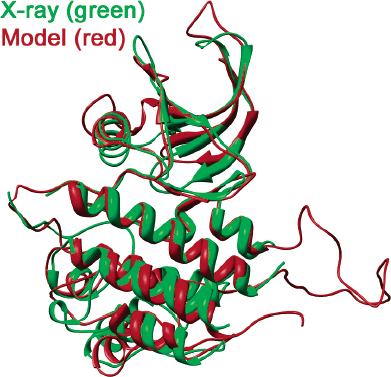
Construction and Validation of a RET TK Catalytic Domain by Homology Modeling. Tuccinardi T, Manetti F, Schenone S, Martinelli A, Botta M. J Chem Inf Mod. 2007 47(2):644-655.